14.2. Sampling Variability#
Goals
Explain sampling variability and why repeated samples differ
Visualize sampling distributions with simulation
Connect standard error to sample size
Recognize law of large numbers behavior
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
sns.set_theme()
We will simulate many samples from a Bernoulli process (think weighted coin with \(p=0.6\) for heads) to see how sample proportions vary.
Key quantities:
True proportion \(p\)
Sample size \(n\)
Sample proportion \(\hat p\)
Sampling distribution: distribution of \(\hat p\) across repeated samples
rng = np.random.default_rng(seed=0)
p_true = 0.6
n = 40
n_sims = 5_000
# Draw many samples and compute sample proportions
samples = rng.binomial(n=n, p=p_true, size=n_sims)
sample_props = samples / n
summary = pd.Series(sample_props).describe(percentiles=[0.025, 0.5, 0.975]).round(3)
summary
count 5000.000
mean 0.600
std 0.078
min 0.350
2.5% 0.450
50% 0.600
97.5% 0.750
max 0.850
dtype: float64
fig, ax = plt.subplots(figsize=(7, 4))
sns.histplot(sample_props, bins=15, stat="density", color="#4c78a8", edgecolor="white", ax=ax)
ax.axvline(sample_props.mean(), color="#f58518", linestyle="--", label=f"mean = {sample_props.mean():.3f}")
ax.axvline(p_true, color="#54a24b", linestyle=":", label="true p")
ax.set(xlabel="sample proportion", ylabel="density", title="Sampling distribution of p-hat (n=40, p=0.6)")
ax.legend()
plt.show()
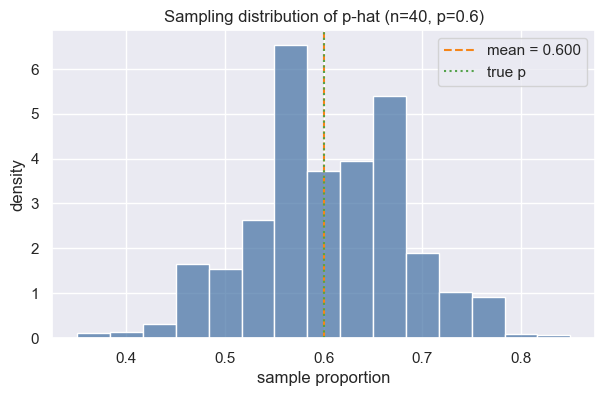
For a sample proportion, the standard error measures the typical deviation of \(\hat p\) from \(p\): $\(\text{SE}(\hat p) = \sqrt{\dfrac{p(1-p)}{n}}\)\( If \)p\( is unknown, we often plug in \)\hat p$.
se_theoretical = np.sqrt(p_true * (1 - p_true) / n)
se_empirical = sample_props.std(ddof=1)
print(f"Theoretical SE: {se_theoretical:.4f}")
print(f"Simulated SE: {se_empirical:.4f}")
Theoretical SE: 0.0775
Simulated SE: 0.0781
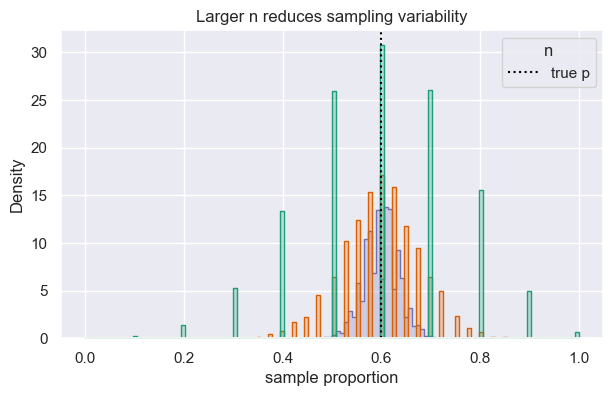
Larger samples shrink variability. Below we compare sampling distributions across different \(n\) while keeping \(p=0.6\) fixed.
def simulate_props(n, p=0.6, sims=5_000):
props = rng.binomial(n=n, p=p, size=sims) / n
return props
ns = [10, 40, 200]
records = []
frames = []
for n_i in ns:
props_i = simulate_props(n=n_i)
frames.append(pd.DataFrame({"n": n_i, "prop": props_i}))
records.append({
"n": n_i,
"mean(p-hat)": props_i.mean(),
"sd(p-hat)": props_i.std(ddof=1),
"SE formula": np.sqrt(p_true * (1 - p_true) / n_i)
})
display(pd.DataFrame(records).round(4))
plot_df = pd.concat(frames, ignore_index=True)
fig, ax = plt.subplots(figsize=(7, 4))
sns.histplot(data=plot_df, x="prop", hue="n", element="step", stat="density", common_norm=False, palette="Dark2", ax=ax)
ax.axvline(p_true, color="black", linestyle=":", label="true p")
ax.set(xlabel="sample proportion", title="Larger n reduces sampling variability")
ax.legend(title="n")
plt.show()
| n | mean(p-hat) | sd(p-hat) | SE formula | |
|---|---|---|---|---|
| 0 | 10 | 0.5994 | 0.1549 | 0.1549 |
| 1 | 40 | 0.5984 | 0.0780 | 0.0775 |
| 2 | 200 | 0.5986 | 0.0348 | 0.0346 |
Takeaways
Repeated samples vary; summarizing their distribution clarifies how much.
Standard error captures typical sampling fluctuation and shrinks as \(n\) grows.
Simulations help check formulas and build intuition about variability.
